Our current research focuses on developing and applying biochemical, molecular biology, and structural biology techniques to understand how RNA structures within viral RNA genomes facilitate the viral life cycle in host cells. This research addresses crucial gaps in the structures and mechanisms of RNA-mediated genome replication and translation in (+) sense RNA viruses, many of which cause human diseases and have caused global pandemics in the past. Our laboratory integrates biochemistry, biophysics, and biology, providing our members with an exceptional opportunity to acquire multiple biochemical and biophysical techniques while conducting advanced RNA structural and molecular biology research. The following are the ongoing research projects in our laboratory.
Structural and mechanistic studies of cap-independent genome translation in (+) strand RNA viruses
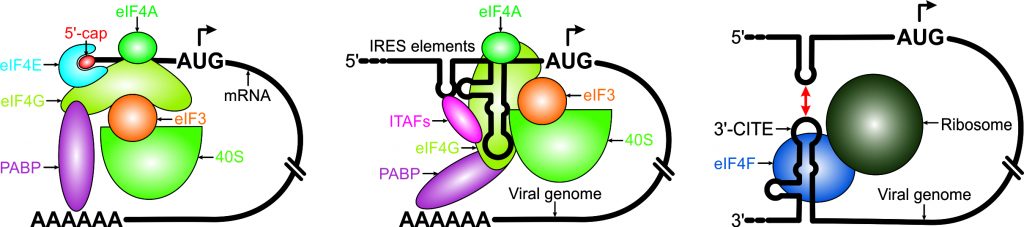
In contrast to the 5′ cap-based canonical translation in most eukaryotes, many viral genomes and a subset of cellular mRNAs are translated via cap-independent mechanisms that involve structured RNA elements such as internal ribosome entry sites (IRESs) and 3′ cap-independent translation elements (3′ CITEs). However, the organization and recognition of these RNA structures by eukaryotic translation initiation factors (eIFs) and ribosomal subunits remain poorly understood. This project investigates how IRESs and 3′ CITEs facilitate cap-independent translation in the genomes of positive-strand RNA viruses. The findings will provide deeper insights into the mechanisms of cap-independent translation initiation in these viruses and may open new avenues for developing RNA-targeted therapeutics against pathogens causing human, animal, and plant diseases.
The crystal structure of saguaro cactus virus (SCV) 3′ translational enhancer that mimics mRNA 5′ cap for eIF4E binding (read the full story: https://www.pnas.org/doi/10.1073/pnas.2313677121)
Structural and mechanistic studies of RNA-mediated enteroviral genome replication
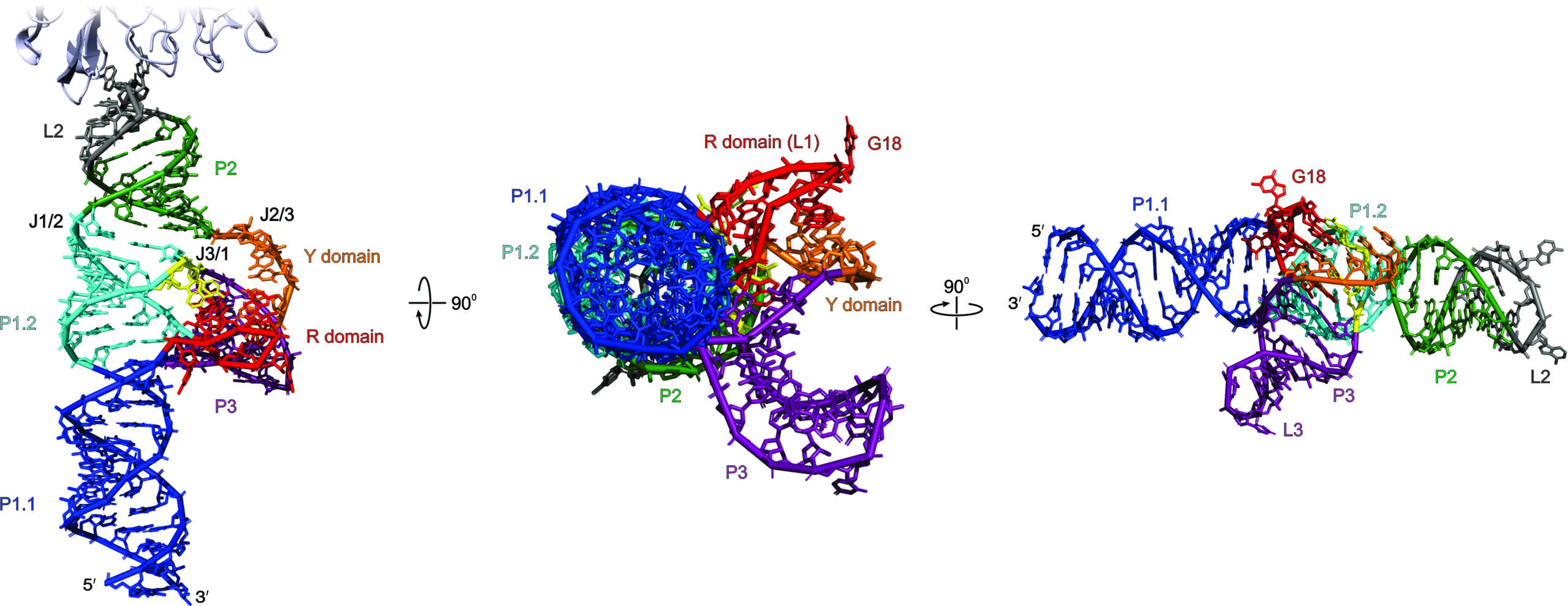
Another major project in our lab focuses on understanding the structural basis of genome replication in enteroviruses. The enterovirus genus of the Picornaviridae family (one of the most prominent families of (+) strand RNA viruses) comprises a class of pathogens that cause a wide variety of human diseases, ranging from the common cold to poliomyelitis, acute flaccid paralysis, and myocarditis. Genome replication in these viruses, which is essential for viral proliferation, occurs in two distinct steps: the synthesis of (-) strand RNA using the genomic RNA as the template, followed by the synthesis of (+) strand genomic RNA using the newly made (-) strand RNA as the template. These processes depend on conserved RNA structures that regulate viral genome replication by recruiting viral and host proteins to form ribonucleoprotein (RNP) complexes and involve these RNPs in virological processes through non-canonical mechanisms. We aim to answer a pressing question regarding replication in enteroviruses: how does an RNA structure-based strategy promote viral genome replication? Our lab’s approach to answering this question relies on determining high-resolution structures of the replication-linked RNAs (REPLRs) found in the viral (+) strand genomes from seven different species of the enterovirus genus using Fab-chaperone-assisted X-ray crystallography, recombinant expression, and purification of viral 3C, 3CD, 3D, 2C, cellular PCBP2, PABP, and hnRNP C proteins; biochemical, biophysical, and structural characterization of the REPLR interactions with these viral and cellular proteins; and finally, determination of high-resolution structures of the REPLR domains found in viral (-) strands and RNP complexes from the same seven enteroviral species.
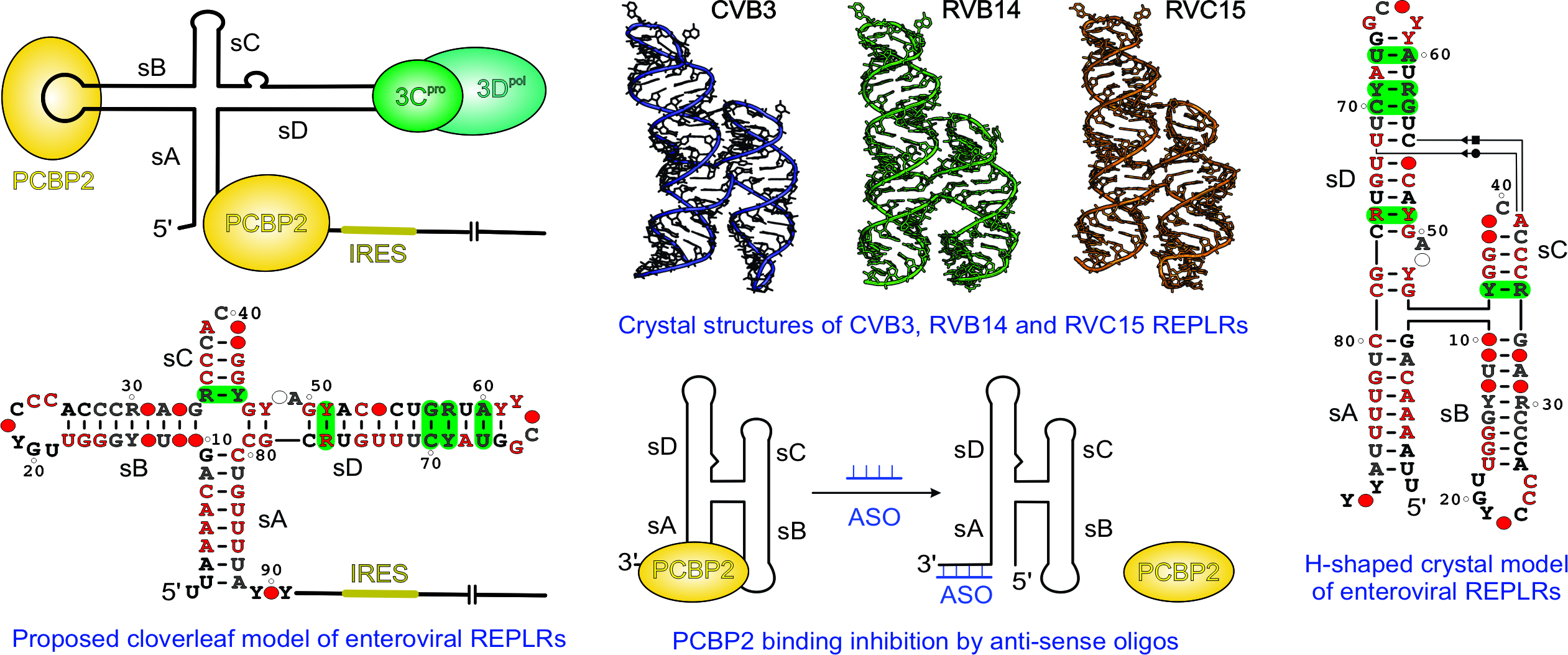
Structural basis for a highly conserved RNA-mediated enteroviral genome replication (read the full story here: https://doi.org/10.1093/nar/gkae627)
Structural studies of HIV-1 RRE and FSE
The primary objective of this project is to gain insights into the structural mechanisms underlying nuclear export of HIV-1 RNA via the Rev Response Element (RRE) and the regulation of HIV-1 protein expression via the Programmed Ribosomal Frame-Shifting Element (FSE). To this end, our lab has used Fab-assisted X-ray crystallography to determine high-resolution crystal structures of the HIV-1 RRE and FSE. Combined with other biophysical and biochemical methods, our lab aims to understand how these RNAs interact with their protein partners to form functional ribonucleoprotein (RNP) complexes.

Mutation-driven RRE stem-loop II conformational change induces HIV-1 nuclear export dysfunction (read the full story here: https://doi.org/10.1093/nar/gkaf583)
Approaches for developing RNA crystallization chaperone modules and novel RNA imaging tools
Among several biophysical methods, X-ray crystallography has been the most widely used and best developed for determining high-resolution structures of biomacromolecules, including RNA molecules. However, RNA structures constitute only a tiny fraction of the high-resolution structures deposited in the Protein Data Bank (PDB), underscoring the challenges of RNA crystallography. We have developed RNA-binding proteins, specifically synthetic antibodies (Fab fragments), as chaperones for RNA crystallization and structure determination. In addition to selecting RNA target-specific Fabs using phage display, we are interested in developing a suite of portable protein-RNA modules via in vitro selection that can be easily engineered into an RNA of interest as affinity tags, with applications in RNA crystallography as well as in RNA imaging and immunoprecipitation.

Synthetic anti-RNA antibody derivatives for RNA visualization in mammalian cells (read the full story here: https://doi.org/10.1093/nar/gkae1275)
The outcomes of our research will provide insights into the mechanisms of fundamental biological processes and unlock ample opportunities to develop targeted therapeutics for genetic and infectious diseases. Moreover, a structural understanding of the landscape of RNA structures and mechanistic insights into how particular structural features enable biological function will be tremendously valuable for developing algorithms to predict new RNA structures using bioinformatics and computational tools.